The ability to visualise host-pathogen interactions across multiple scales, from individual molecules to whole organisms, unlocks unprecedented opportunities for breakthroughs in our basic understanding of infectious disease biology. However, working with the live human or animal pathogens also brings unique challenges. By sharing experiences, skills and resources, researchers in Cambridge are redefining what is possible in infections disease imaging and illuminating novel biological processes that can be targeted for the development of next-generation therapies.
Events and Progress
Collaborative workshop
To explore challenges, future technologies and collaboration opportunities in the field of infection imaging at Cambridge, members of the Infection and Immunity Research Theme from across 6 Departments and Initiatives met for a collaborative workshop in March 2022. Five speakers presented their imaging research and techniques, followed by a productive discussion to identify the barriers to infection and immunity imaging research and explore new imaging modalities that could be pursued to drive progress in future.
As a result of the meeting, researchers are coordinating an application for a cryo-microscope and plunge-freezing equipment so that infected samples can be prepared here in Cambridge for ultrastructural imaging analysis (such as that described in the recent publication below). They are also coordinating with existing microscopy facilities to maximise their availability for pathogen research under appropriate biosafety containment conditions.
Investments in imaging
Enhancing the capacity within the School of Biological Sciences to image infected cells will facilitate important progress and understanding across a range of diseases, from viruses that cause childhood illnesses (Zika virus and astroviruses) or highly prevalent lifelong infections (herpes), to pathogenic bacteria (Pseudomonas and Chlamydia spp) and the Plasmodium parasites that cause malaria. We are pleased to have recently secured funding for a cutting-edge automated microscope (Incucyte SX5) that allows us to monitor infected cells over the course of days via brightfield and three-colour fluorescent imaging. Installed in August 2022, the microscope will be housed in the biosafety containment (CL2) laboratory in the Division of Virology.
“This new microscope is cutting-edge technology that will allow us to closely monitor large numbers of infected cells for days at a time. Having this capacity in Cambridge will hugely advance our knowledge of pathogen-host interactions and infection dynamics, illuminating real-time changes in cell morphology and behaviour that are caused by infection.”, said Valeria Lulla, group leader from the Department of Pathology who coordinated the purchase.
Research Highlight
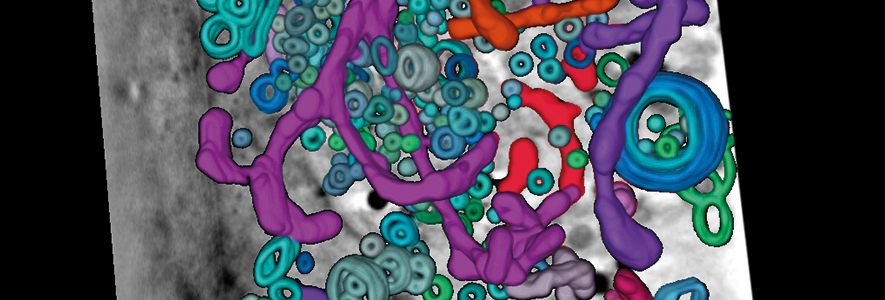
Theme members in the Department of Pathology have used an advanced imaging technique called cryo-soft-X-ray tomography to see how viruses disrupt and damage the vital internal structures of our cells. Importantly, this technique allows researchers to obtain a high-resolution 3D view of the cell’s internal structure without the need for any complicated sample manipulation or staining.
Working collaboratively with colleagues in the Department of Chemical Engineering and with collaborators at Diamond Light Source beamline B24, the researchers studied how infection by herpes simplex virus alters the shape and arrangement of internal structures within the cell. They saw that infection dramatically changes the architecture of mitochondria—the powerhouses of the cell. It also causes accumulation of the small membrane-bound compartments (vesicles) that act as a biological “shipping service” to shuttle components around the cell, suggesting that the virus is re-routing this cargo.
“Under normal, healthy conditions, the mitochondria in cells are a mixture of shapes and lengths. When we infected the cells with the herpes simplex virus, we saw a big shift towards long and highly-branched mitochondria. This is likely to effect on the cell’s ability to make energy, and it may impair the function of mitochondria as molecular ‘trip-wires’ to detect virus infection.” says Stephen Graham, senior author of the paper. “Herpes simplex virus and humans have evolved together – the virus knows our cells inside out and is exceptionally good at modifying them to suit its own ends. Using this latest imaging technology, we can peer inside cells to understand exactly what changes the virus is making. This gives us a starting point to arrest those changes, and ultimately to stop the virus in its tracks”.
Vesicles (rings) and mitochondria (tubes) in a herpes simplex virus infected cell, visualised using cryo-soft-X-ray tomography.
Publication details: Near-native state imaging by cryo-soft-X-ray tomography reveals remodelling of multiple cellular organelles during HSV-1 infection. Nahas KL, Connor V, Scherer KM, Kaminski CF, Harkiolaki M, Crump CM, et al. PLoS Pathog 18(7): e1010629. doi.org/10.1371/journal.ppat.1010629
Key Collaborators
Katerina Artavanis-Tsakonas, Pathology
Andrew Blagborough, Pathology
Clare Bryant, Veterinary Medicine
Betty Chung, Pathology
Colin Crump, Pathology
John Doorbar, Pathology
James Edgar, Pathology
Jenny Gallop, Biochemistry and Gurdon Institute
Stephen Graham, Pathology
Richard Hayward, Pathology
Nerea Irigoyen, Pathology
Ben Luisi, Biochemistry
Valeria Lulla, Pathology
Ioanna Mela, Pharmacology
Vito Mennella, MRC Toxicology Unit
Catherine Merrick, Pathology
Cahir O'Kane, Genetics
Emma Rawlins, Physiology, Development and Neuroscience and Gurdon Institute
Olivier Restif, Veterinary Medicine
Jeane Salje, Pathology and Biochemistry
Work with us
We welcome opportunities to collaborate with industry partners, policy makers and academics. If you are interested in working with us, please contact Dr Abi Herrmann, Research Strategy Facilitator.